
OncoPPIMiner is a simple-to-use database resource that allows the search for proteins frequently involved in edgetic perturbations (herein termed oncoproteins) at either the cancer-type level or at the cancer-subtype specific level. Additionally, we provide experimental evidence from literature sources indicating the specific roles the oncoproteins play in tumorigenesis. Our research is enhanced due to the recent advances in next-generation sequencing technologies that have enabled comprehensive cancer genomic testing by molecular pathologists across multiple tumor types. Apart from somatic mutations in the cancer driver genes (i.e significantly mutated genes - SMGs), isoform switching (IS) is another recently characterized hallmark of cancer. IS often translates to the loss or gain of domains responsible for mediating protein-protein interactions and thus, the re-wiring of the interactome. While there is a multitude of databases providing information about cancer, there are no elaborate and large scale databases for prominent proteins (or biomarkers) frequently involved in edgetic perturbations in cancer. We have created a literature-based annotation resource of cancer biomarkers at the PPIN level with potentially actionable proteins for translational oncologists and clinicians to facilitate experimental research in the continued quest for druggable proteins at the protein-protein interaction network level.
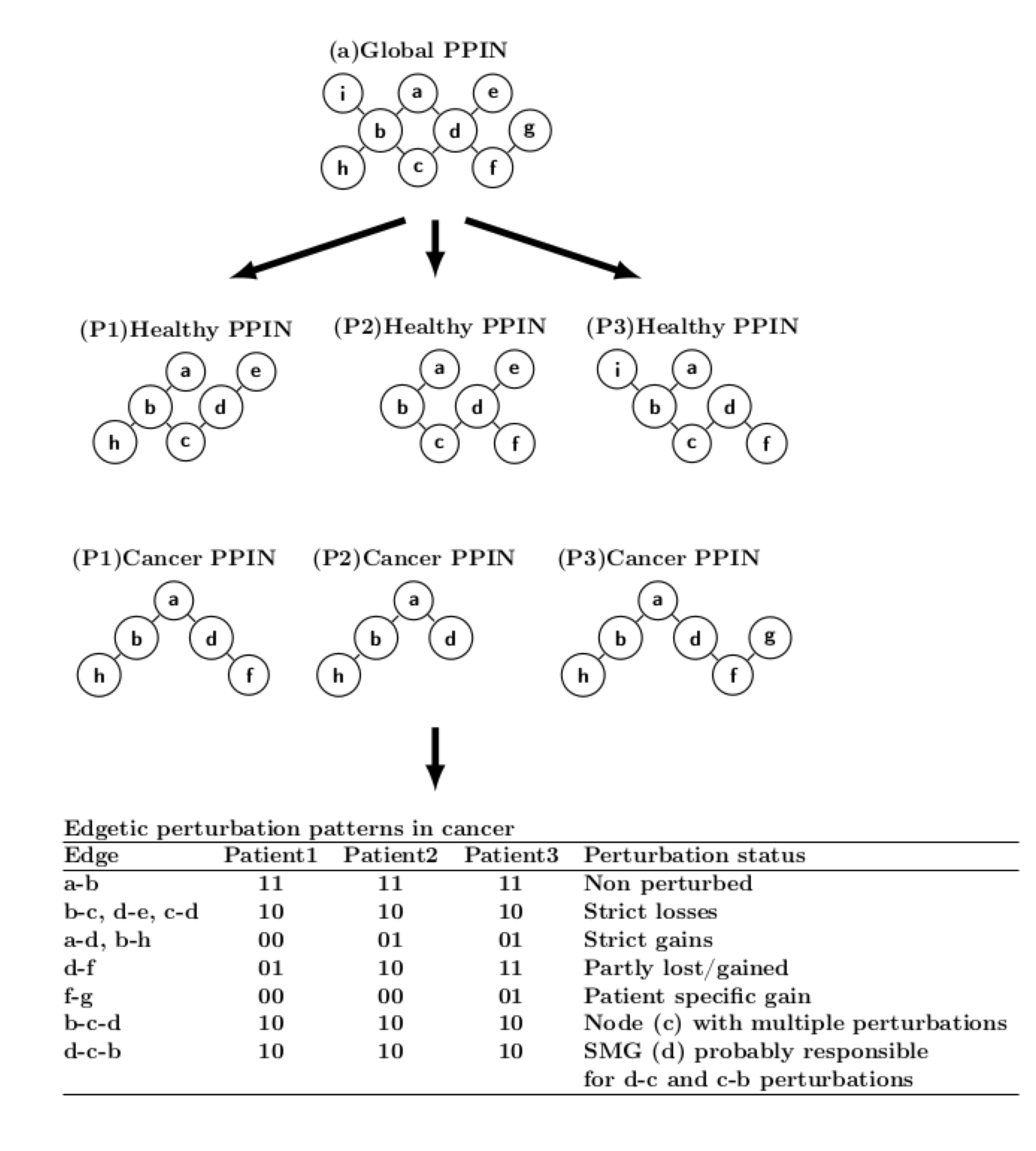
To find prominent proteins frequently involved in edgetic perturbations at the multi-cancer, cancer type,
and cancer subtype levels, proteins were ranked according to
the number of perturbations they and their first network neighbors are involved in. Perturbations
associated with the first neighbors were only counted if the protein itself was associated with a perturbation.
For instance, perturbation of edges b-c, c-d and d-e would give a rank of 3 for node c, and a rank of 1
each for nodes b, d and e. Node c would thus be considered as significantly perturbed across all patient samples.
Using the gene names for each of the above proteins, we then searched Google Scholar and PubMed for experimental evidence
linking them to cancer development.
Click to download all genes expressed per cancer type for paired healthy and cancer samples (Dataset 1)
DownloadClick to download the sampled edgetic perturbations in BLCA and BRCA after network randomization(Dataset 2)
DownloadClick to download all the edgetic perturbations in all the patients of a cancer type (Dataset 3)
DownloadClick to download the reproducible edgetic perturbations in patients of a cancer type (Dataset 4)
DownloadClick to download additional analyses results (e.g KEGG, GO enrichment, disease-gene relations) in Table format (Dataset 5)
DownloadThis resource is the joint work between the Dmitrij Frishman Lab at The Technical University of Munich and Dr. Goar Frischmann, a Biocurator at the HelmholtzZentrum München.